The Central Dogma of Biology

How did our physical features come about? What tells our body to repair that wound? To examine things further, we will dive into how cells are able to send genetic information to where they need to be and understand the mechanisms that facilitate them.
Click below to go to the main reviewers:
Table of Contents
- A Brief History of DNA and Its Role in Genetics
- DNA Replication – It Follows Directions
- Gene Expression – From DNA to RNA to Protein
- Mutations – Not as Drastic as Those of X-Men
- The Genetics of Viruses and Bacteria
- References
- Download Article in PDF Format
- Test Yourself!
A Brief History of DNA and Its Role in Genetics
We know that DNA holds the information for characters that are inherited, but how did it come to be and what’s the mechanism behind it?
The discovery of DNA’s genetic role can be traced back to medical officer Frederick Griffith who was trying to develop a vaccine against pneumonia. He was observing two strains (or varieties) of the pneumonia bacteria: one that is harmless and the other causes the disease in mammals (it is pathogenic). He killed the pathogenic ones and mixed the remains with the harmless strain and was surprised that some of the living strains became pathogenic.
Furthermore, descendants of the transformed bacteria also acquired the ability to cause the disease.
A component of the dead bacteria caused this ability to be inherited. It sparked the race to understand why, and it was cemented later when biologists Alfred Hershey and Martha Chase experimented and showed that DNA is the genetic material, specifically of the bacteriophage (or simply phage), viruses that exclusively infect bacteria.
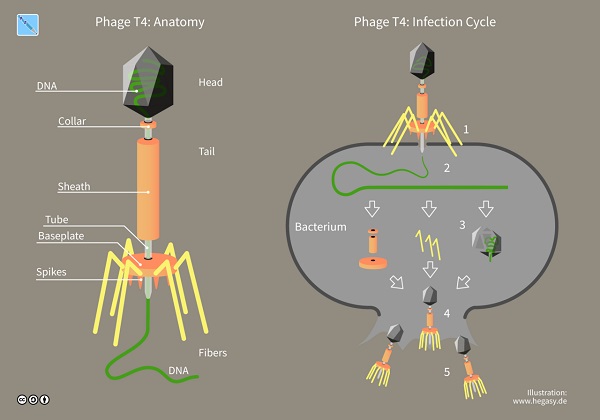
Once biologists had confirmed that DNA was the molecule for heredity, they began studying its structure. As discussed previously, DNA and RNA are polymers of nucleotides, and their chemical components contribute to their form (i.e., that the sugar-phosphate backbone is towards the outside of the strand and forces the nitrogenous bases to swivel inwards of the molecule) and their specific function in gene expression.

How the shape of DNA as a double-stranded helix was known can be traced to three people: Rosalind Franklin, James Watson, and Francis Crick.
Rosalind Franklin was able to take an X-ray image of the DNA, and the two men expanded on it. Thus, we were able to understand the chemical components in addition to the DNA’s structure and how this contributes to DNA replication
DNA Replication – It Follows Directions
Watson and Crick proposed that the specific pairing of the DNA’s bases accounts for its ability to be copied. Their model predicted that when a double helix replicates, each of the resulting strands will have one old strand from the parental molecule and another newly created strand. This model came to be known as the semiconservative model because half of the parental strand is maintained in the daughter strands (hence it is conserved).
In concept the process is simple, but in cells, it requires the coordination of many enzymes and other proteins.

Replication of DNA begins at particular sites called origins of replication, short stretches of specific DNA sequences. Proteins attach at the sites to separate the strands of the helix and replication ensues in both directions, creating replication “bubbles.”
Imagine the DNA helix as two parallel lines; proteins will attach at different points of that line, opposite each other, and separate the lines at those points creating a bubble.
As the bubbles grow while separating the lines, new lines will get created until such time that the original parallel lines get separated but will be paired with new lines. In terms of chemical composition, a DNA strand has two ends: the 3’ (“three-prime”) end and the 5’ (“five-prime”) end which refers to the carbon atom of the nucleotide sugar. The 3’ carbon atom is attached to an -OH group while the 5’ carbon is attached to the phosphate group.
Each DNA strand runs in the opposite direction to the other, and this contributes to DNA replication, offering directionality.

Because molecules are reactive, when DNA strands are separated, they must be stabilized when linking to a growing daughter strand occurs. DNA polymerases are designed to facilitate this, and these proteins add the daughter strands only to the 3’ end of the strand. As a consequence, the parent strand having a 3’ to 5’ direction will have continuous replication while the opposite strand gets replicated in short pieces and linked together. These short pieces are called Okazaki fragments and the protein that links them with each other to form a continuous daughter strand is called DNA ligase.
In addition to adding sequences to the DNA chain, DNA polymerase also proofreads to remove bases paired incorrectly during replication. Together with DNA ligase, the two proteins also repair DNA damaged by radiation or other toxic chemicals from the environment.
We have been introduced to DNA and how it replicates. Next, we will look at how DNA becomes a template for the characteristics and features we all see from the different life forms around us.
Gene Expression – From DNA to RNA to Protein
We can now define genotype and phenotype based on the structure and function of DNA.
Genotype is an organism’s genetic makeup and is the heritable information stored in the nucleotide sequences of DNA. Phenotype is the organism’s physical traits. So, what connects the two exactly?

Well, we know that DNA specifies traits by dictating the synthesis of proteins. In other words, proteins are the link between genotype and phenotype.
Genes alone do not build proteins directly. This fundamental concept is referred to as the “central dogma of biology” which refers to the flow of genetic information and its expression. This dogma tells us that the molecular chain of command is from the DNA in the cell, to RNA, to protein synthesis in the cytoplasm.
The two main stages are transcription, the synthesis of RNA based on instructions from DNA; and translation, the synthesis of protein under the direction of RNA.
1. Transcription in Prokaryotes
The transcription process in prokaryotes is simpler compared to eukaryotes. Once the DNA strands are separated, one strand serves as a template for a new RNA molecule, and the other is unused.
The enzyme for transcription is RNA polymerase which moves along the gene, following the base-pairing rules. A specific sequence called promoter acts as a binding site for RNA polymerase and determines where transcription starts. RNA polymerase adds to the chain until it reaches a sequence of DNA bases called the terminator, which signals the end of the gene.
The transcription of the gene in prokaryotes is composed of three major steps:
- Initiation – which involves the attachment of RNA polymerase to the promoter to start RNA synthesis.
- Elongation – the newly formed RNA strand grows with the growing RNA peeling away from its DNA template. This allows the separated DNA strands to come back together in the region already transcribed
- Termination – When RNA polymerase reaches the terminator DNA, the polymerase molecule detaches from the newly formed RNA strand and the gene.

2. Gene Expression in Eukaryotes
Genes provide instructions for making specific proteins. The bridge linking DNA and protein synthesis is the nucleic acid RNA.
DNA is transcribed into RNA before it is translated into proteins. Transcription and translation are linguistic terms but are helpful in explaining the process.
When DNA forms RNA, the process is called transcription, and think of it as if the language of DNA has been rewritten as a sequence of RNA. Remember that the nitrogenous base T (thymine) of DNA is substituted for U (uracil) in the latter. This is because RNA was synthesized using DNA as the template.
During translation, there is a shift in language from the nucleotide sequence of RNA to the amino acid sequence of the polypeptide. Recall that there are only four nucleotides in RNA, but they must somehow specify the 20 amino acids. If one nucleotide only corresponds to one amino acid, this will only account for 4 amino acids.
But what if two letters are needed to make one acid? What if there are more?

Experiments have verified that the flow of information from gene to protein is based on a triplet code: that the instructions for the amino acid sequence of a polypeptide are a series of nonoverlapping three-base “words” called codons.
With three base units accounting for all 20 amino acids, scientists were able to crack the genetic code, and among the codons, they observed the following:
- AUG has two functions: (1) it codes for the amino acid methionine (Met) and (2) provides the signal for starting the polypeptide chain. You can say it is the start codon.
- Among the codons, UAA, UGA, and UAG do not code for amino acids because they mark the end of translation (thus, being referred to as stop codons).
RNA is structurally different from DNA. For this reason, RNA also has some specific functions that DNA cannot do. These functions manifest as different RNA types also, which we will look into next.
a. Types of RNA and their Functions

i. Messenger RNA – Delivering Instructions from the DNA
The kind of RNA that encodes for amino acid sequences is called messenger RNA (mRNA) because it conveys instructions from DNA to the translation mechanism of the cell. mRNA is transcribed from DNA with the information used to translate it into polypeptides.
In prokaryotic cells without a nucleus, transcription and translation occur in the cytoplasm. In eukaryotic cells, however, the mRNA must first exit the nucleus via nuclear pores and enter the cytoplasm where proteins wait for instructions for translation.
Eukaryotes modify and process mRNA before they leave the nucleus. One way is adding additional sequences on the ends of the RNA template as either a small cap at the 5’ end or a long tail at the 3’ end. These facilitate the export of mRNA from the nucleus and protect mRNA from degradation.
Another kind of RNA processing is necessary because most of the sequences in the RNA interrupt the nucleotides that code for amino acids or they don’t code for anything. It’s like nonsense words were put randomly within a story. These internal non-coding regions are called introns and must be removed. The remaining sequences, the expressed parts of the gene called exons, are then pasted together.
This cut and paste mechanism is called RNA splicing and ensures no error in the process occurs.
ii. Transfer RNA – Picker of Proteins
Translation involves more elaborate mechanisms than transcription but requires the processed mRNA. Once present, other enzymes and energy are used to translate this mRNA into proteins.
Like translation in the real world, an interpreter is required to convert the language of mRNA into proteins.
This interpreter within the cell is a special type of RNA called transfer RNA (tRNA), and it transfers amino acids from the cytoplasm into a growing polypeptide chain in a ribosome. To perform the task, it carries out two functions: (1) pick up the appropriate amino acids and (2) recognize the appropriate codons in mRNA. As usual, the structure contributes to its unique function.
The tRNA contains a special site composed of triplet bases called an anticodon. This site complements and recognizes a codon triplet on mRNA due to base pairing.
On the opposite end of the tRNA molecule is the site where one specific kind of amino acid attaches. Different tRNA molecules occur for different amino acids, and each also has specific enzymes that assist in joining the correct tRNA to its corresponding amino acid, with the use of ATP.
iii. Ribosomal RNA – Facilitator of Translation
The final components of the translation process are the ribosomes, structures in the cytoplasm that coordinate both mRNA and tRNA and catalyze the production of polypeptides.
Ribosomes are composed of two subunits – a large and small subunit, each made up of proteins and a kind of RNA called ribosomal RNA (rRNA).
Ribosomes of bacteria and eukaryotes perform similarly, but those of eukaryotes are slightly larger. These slight differences are significant in medicine and allow for specific targeting of pathogenic bacteria without harming ribosomes in the body such as when using the drug streptomycin.
We took a closer look at the different types of RNA and their functions. Let’s return and continue the process of utilizing these different types of RNA.
3. Translation
Like transcription, translation can also be divided into three phases: initiation, elongation, and termination.
Initiation occurs in two steps:
- An mRNA molecule binds to a small ribosomal subunit. A special initiator tRNA base pairs with the start codon, which helps start up the translation. The initiator tRNA carries the amino acid methionine (Met) and its anticodon, UAC, pairs with the start codon AUG.
- The large subunit then binds to the small subunit, creating a functional ribosome that has two sites – the P site where the initiator tRNA binds to and which holds the growing polypeptide, and the A site which is ready for the next amino acid-bearing tRNA.

Elongation is a three-step process:
- The anticodon of incoming tRNA pairs with the mRNA on the A site.
- The polypeptide separates from the tRNA in the P site and attaches to the amino acid carried by the tRNA in the A site; with ribosomes catalyzing the formation of peptide bonds. A tunnel in the ribosome allows for the growing chain a way out of the molecule.
- The P site tRNA leaves the ribosome, and the A site’s tRNA moves to P. Since the tRNA is bonded to mRNA, when tRNA moves the mRNA also moves. This causes a new tRNA to bind to the A site. This elongation process repeats until a stop codon reaches a ribosome’s A site. This also marks the termination stage, and the completed polypeptide is freed from the last tRNA. The ribosome then splits back into its separate subunits.
Mutations – Not as Drastic as Those of X-Men
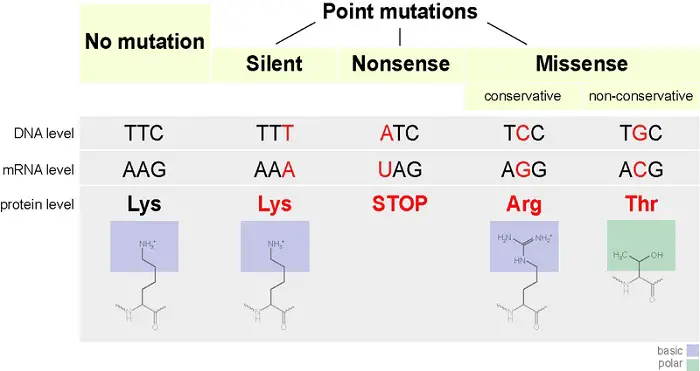
Although the molecular process of gene expression is rigorous, complex, and highly regulated, it is not infallible. Any change to the genetic information of a cell is called a mutation, and there are several types of mutations.
A nucleotide substitution occurs when one nucleotide in mRNA gets replaced. This results in three scenarios:
- When the substituted nucleotide still encodes for the same amino acid, this will have no effect at all leading to a silent mutation.
- The substituted nucleotide causes a change in the amino acid sequence. This is a missense mutation.
- When the substitution causes the codon to be one of the stop codons, it is called nonsense mutation which results in a prematurely terminated protein that may not function properly.
Because mRNA is read as a series of nucleotide codons during translation, the addition or subtraction of nucleotides may alter the resulting sequence of proteins. Such a mutation is called a frameshift mutation and occurs whenever the mRNA gets inserted or deleted by a nucleotide that is not a multiple of three, resulting in the nucleotide sequences regrouping into different codons.
Consider the following sentence:
“The cat ate the rat”
If we remove c, the result is a nonsensical message: “The ata tet her at.”
Frameshift mutations in a way will most likely produce nonfunctional polypeptides and often have disastrous effects. Mutations can arise in many ways: from errors during DNA replication or recombination; or due to effects of physical or chemical agents called mutagens.
The Genetics of Viruses and Bacteria
Bacteria and viruses served as models in uncovering the molecular mechanisms behind heredity. The latter is still debated by biologists whether it should be considered a living organism or not.
In a sense, viruses are infectious particles consisting of nucleic acids wrapped in a protein coat called a capsid and in some, a membranous envelope. Unlike other organisms, viral genomes can be DNA or RNA and usually consist of a single molecule of nucleic acid, which may be linear or circular.
Viruses are parasites that reproduce once they are inside cells. Viruses take two routes to make use of host cells: the lytic cycle and the lysogenic cycle. Both cycles begin when the phage enters the bacterium and forms a loop.

In the lytic cycle, the virus turns the cell into a virus-producing factory, and the cell soon lyses (or bursts) as it releases the viral products which proceed to infect other cells. In the lysogenic cycle, after the viral DNA gets inserted into the bacterial chromosome (referred to as a prophage), every time the bacterial cell divides, it replicates the viral DNA and passes copies of it onto daughter cells.
The lysogenic cycle allows the virus to spread without killing the host cells they depend on. The virus which causes the COVID-19 disease is a lytic virus targeting the alveoli in the lungs hence the symptoms associated with the respiratory system.
Bacteria are also valuable in genetics research, being representatives of prokaryotes. Most of a bacterium’s DNA is found in a single chromosome, a closed loop of DNA with associated proteins. Bacterial cells reproduce by replication of the bacterial chromosome followed by binary fission.
Although binary fission is an asexual reproductive process, bacteria have ways to produce new combinations of genes:
- In transformation, the bacteria take foreign DNA directly from the surrounding environment.
- The second means, which brings together genes of different bacteria, is using bacteriophages in a process called transduction.
- The last is through swapping genes between some bacterial cells, whether they are the same or of different species, through the physical process called conjugation. The donor cell has hollow appendages called sex pili which attach to the recipient cell and pull the two cells together, like a grappling hook. The donor cell replicates its DNA as it transfers it, so the recipient cell doesn’t end up lacking any genes. The ability of a donor cell to carry out conjugation is based on a specific piece of DNA called the F factor (F for fertility).
References
Alberts, B., Johnson, A., Lewis, J., Raff, M. C., Roberts, K., Walter, P., … Hunt, T. (2008). Molecular Biology of The Cell. New York: Garland Science.
Armstrong, P. (2007). All things Darwin: An encyclopedia of Darwin’s world. Westport, Connecticut: Greenwood.
Campbell, N. A., Cain, M. L., Minorsky, P. V., Reece, J. B., Urry, L. A., & Wasserman, S. A. (2018). Biology: A global approach. Harlow, Essex, England: Pearson Education Limited.
Chiras, D. D. (2008). Human Biology. Sudbury, Mass: Jones and Bartlett Publishers.
Fowler, S., Roush, R., & Wise, J. (2013). Concepts of Biology. Houston, Texas: OpenStax. Available at https://openstax.org/books/concepts-biology/pages/1-introduction.
Hine, R. (2019). A Dictionary of Biology. Oxford, US: Oxford University Press.
Hillis, D. M., Moritz, C., & Mable, B. K. (1996). Molecular systematics. Sunderland, Mass: Sinauer.
Hollingsworth, P. M., Bateman, R. M., Gornall, R. J., & Conference on Advances in Plant Molecular Systematics. (1999). Molecular systematics and plant evolution. New York: Taylor & Francis.
Jamora, C., Fuchs, E. (2002). Intercellular adhesion, signaling, and the cytoskeleton. In: Nature Cell Biology. Volume 4, E101–E108. Available at https://doi.org/10.1038/ncb0402-e101
Karp, G., Iwasa, J., & Marshall, W. (2018). Cell biology. Hoboken, NJ: John Wiley.
Lecuit, T., Pilot, F. (2003). Developmental control of cell morphogenesis: a focus on membrane growth. In: Nature Cell Biology. Volume 5, pages 103–108. Available at https://doi.org/10.1038/ncb0203-103
Lodish, H. F. (2013). Molecular Cell Biology. New York: W.H. Freeman and Co.
Meerman, R., & Brown, A. (2014). When somebody loses weight, where does the fat go? British Medical Journal. Vol. 2014. Issue 349.
Nelson, D. L., & Cox, M. M. (2017). Lehninger Principles of Biochemistry (7th ed.). W.H. Freeman.
Ochman, H. (2016). Microbial evolution: A subject collection from Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Reece, J. B., & Campbell, N. A. (2011). Campbell Biology. Boston: Benjamin Cummings/Pearson.
Simpson, M. G. (2019). Plant systematics. Amsterdam Academic Press.
Taylor, M. R., Simon, E. J., Dickey, J., Hogan, K. A., & Reece, J. B. (2018). Campbell Biology: Concepts & connections. NY: Pearson.
Next topic: Biological Diversity
Previous topic: Cellular Metabolism
Return to the main article: The Ultimate Biology Reviewer
Download Article in PDF Format
Test Yourself!
1. Practice Questions [PDF Download]
2. Answer Key [PDF Download]
Written by Earl Jeroh Bacabac
in College Entrance Exam, LET, NMAT, Reviewers, UPCAT
Earl Jeroh Bacabac
Earl’s love for the sea fueled his goal to become a marine biologist. He obtained his Bachelor’s Degree in Biology from the University of the Philippines Visayas while also being a DOST scholar. His passion for the marine environment is rivaled by his diverse interests in music, the arts, and video games.
Copyright Notice
All materials contained on this site are protected by the Republic of the Philippines copyright law and may not be reproduced, distributed, transmitted, displayed, published, or broadcast without the prior written permission of filipiknow.net or in the case of third party materials, the owner of that content. You may not alter or remove any trademark, copyright, or other notice from copies of the content. Be warned that we have already reported and helped terminate several websites and YouTube channels for blatantly stealing our content. If you wish to use filipiknow.net content for commercial purposes, such as for content syndication, etc., please contact us at legal(at)filipiknow(dot)net